Team members
Thomas Rival
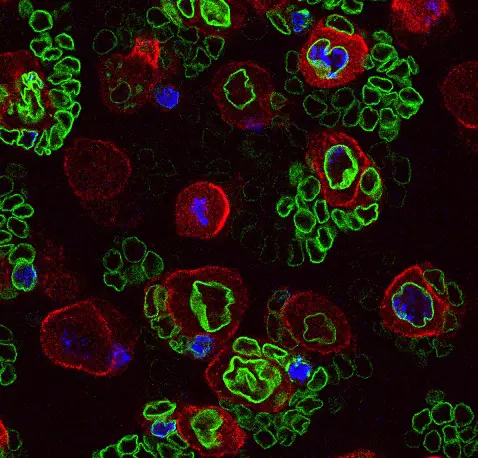
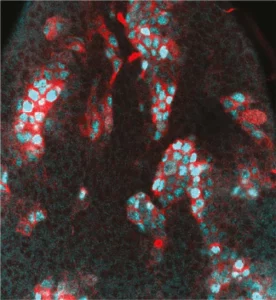
Neural stem cell plasticity
Our team deciphers the mechanisms that control stem cell proliferation during development, childhood cancers and tissue regeneration. We mainly investigate these processes in the central nervous system.
When we will fully understand how stem cells are controlled in our bodies, will we be able to regenerate injured organs or cure cancer?
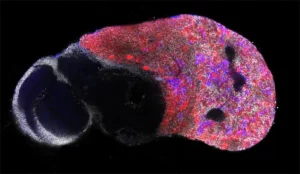
Our laboratory combines two of the most powerful model organisms (Drosophila and the avian embryo) with new technologies to manipulate and study neural stem cells with unprecedented molecular precision. This enables us to decipher the different levels of regulation controlling the unfolding of genetic programs in stem cells during normal development, regeneration and in cancers with developmental origins such as pediatric cancers.
We study how stem cells are controlled by epigenetics, transcription and translation, but also by biochemical and mechanical signals produced by their environment.
Join us if you want to contribute too!
Publications
Regulation of developmental hierarchy in Drosophila neural stem cell tumors by COMPASS and Polycomb complexes
Coopted temporal patterning governs cellular hierarchy, heterogeneity and metabolism in Drosophila neuroblast tumors
Developmental regulation of regenerative potential in Drosophila by ecdysone through a bistable loop of ZBTB transcription factors
Neural stem cell-encoded temporal patterning delineates an early window of malignant susceptibility in Drosophila
Protection of neuronal diversity at the expense of neuronal numbers during nutrient restriction in the Drosophila visual system
Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila
Serotonin signaling regulates actomyosin contractility during morphogenesis in evolutionarily divergent lineages
Regulation of developmental hierarchy in Drosophila neural stem cell tumors by COMPASS and Polycomb complexes
Temporal patterning in neural progenitors: from Drosophila development to childhood cancers
Coopted temporal patterning governs cellular hierarchy, heterogeneity and metabolism in Drosophila neuroblast tumors
Developmental regulation of regenerative potential in Drosophila by ecdysone through a bistable loop of ZBTB transcription factors
Two distinct mechanisms silence chinmo in Drosophila neuroblasts and neuroepithelial cells to limit their self-renewal
Neural stem cell-encoded temporal patterning delineates an early window of malignant susceptibility in Drosophila
Building a brain under nutritional restriction: insights on sparing and plasticity from Drosophila studies.
Protection of neuronal diversity at the expense of neuronal numbers during nutrient restriction in the Drosophila visual system
Temporal specification of neural stem cells: insights from Drosophila neuroblasts.
Temporal control of neuronal diversity: common regulatory principles in insects and vertebrates?
Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila
Signaling meets chromatin during tissue regeneration in Drosophila.
Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs.
Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila.
Spatial and temporal control of transgene expression in vivo using a heat-sensitive promoter and MRI-guided focused ultrasound.
A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development.
News
Watch how model organisms are used at the IBDM to investigate normal and derailed stem cell trajectories.
Join the IBDM for your internship!
Seeking for your Master internship? The IBDM seems like the right place to do it? Check out our offers.
Youth & Science: Delta Festival
Congratulations to Robert Kelly, Frank Schnorrer, Cédric Maurange, Bianca Habermann and Delphine Delacour!
3 motivated and talented students successfully defended their thesis between February and July 2023.
IBDM Marseille inspires young minds: engaging primary school children on childhood cancer (“Contre le cancer, j’apporte ma pierre”) and interacting with high school students through immersive experiences (DECLICS).
Several awards for IBDM members !
Epigenetic factors regulate tumor hierarchy
7 IBDM teams have received grants from ANR
7 IBDM teams have received grants from the Agence Nationale pour la Recherche (ANR) in 2021. Congratulations to Vincent Bertrand, Harold Cremer, Pascale Durbec, André Le Bivic, Pierre-François
In a recent work published in eLife, Cédric Maurange’s team identifies a genetic program that organizes the metabolic heterogeneity of cells in a pediatric neural tumor model.
During this M2 project, the student will work on a recently developed model of medulloblastoma in the chick embryo.
We welcome applications from highly motivated, curiosity-driven PhD candidates, interested in the topic of Developmental Neurobiology, to join Cédric Maurange’s team. The position is funded for 3 years. Research in