Animal species are characterized by a great diversity of morphological forms, color patterns and behaviors. Our research aims to understand how these different traits are encoded by the genetic information contained in the genome, and to elucidate how these traits evolve between species. To do this, we are studying different species of insects, mainly Drosophila flies but also ladybirds. We are studying the formation and evolution of color patterns on the wings of these insects, the morphogenesis of certain body parts, and reproductive behaviors such as courtship or the choice of egg-laying site.
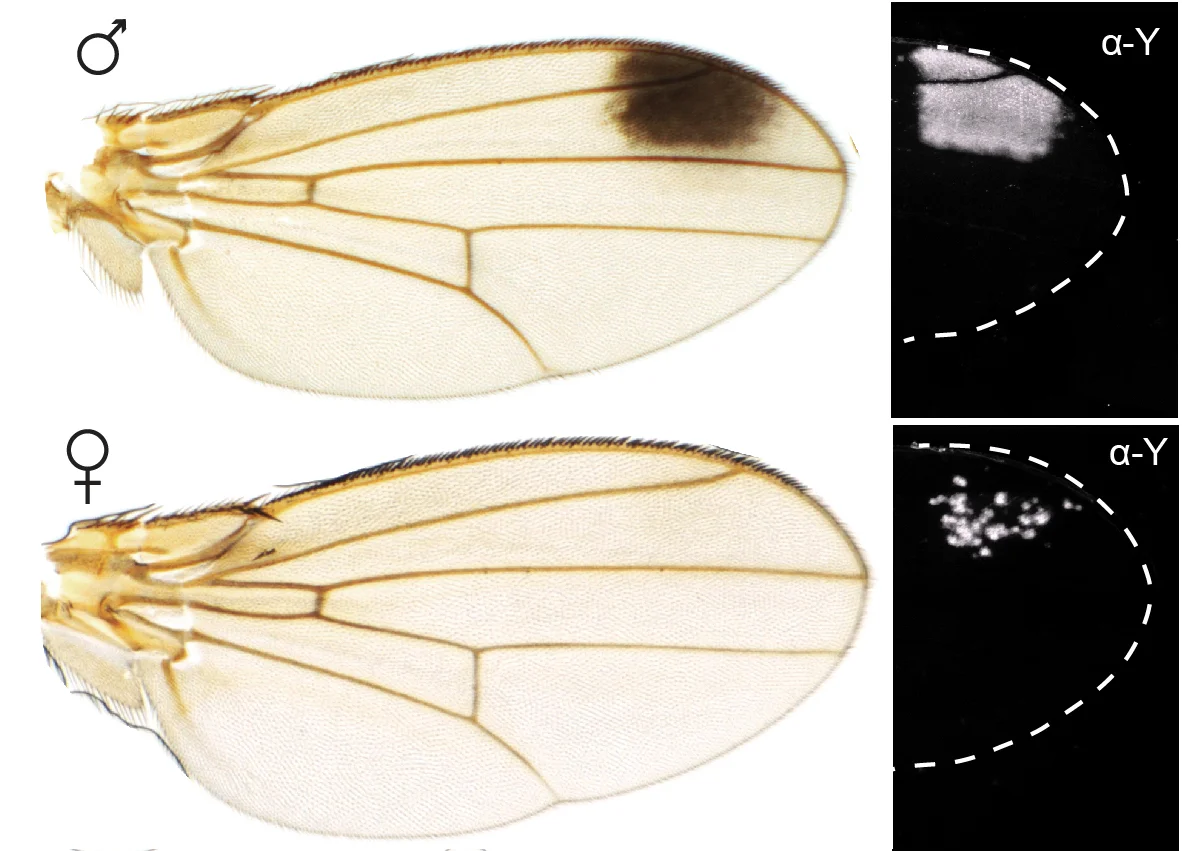
We are studying in particular a Drosophila species native to Asia and recently arrived in Europe, Drosophila suzukii. This fly is an agricultural pest that targets many crop fruit while they are still ripening, such as strawberries, cherries, raspberries and many others, causing very important economic losses. Drosophila suzukii behaves unlike most other Drosophila species, which prefer to lay their eggs on decaying fruit. We seek to understand why and how the choice of egg-laying site has changed in Drosophila suzukii. We are studying the genetic mechanisms that control oviposition behavior, but also the formation of the oviposition organ (which has lengthened in Drosophila suzukii compared to other species, allowing it to pierce the skin of ripe fruits). Finally, this species, like some evolutionary close cousins, is recognizable by the presence of a small black spot at the wing tips, but only in males. We are trying to understand how this coloration pattern appeared on the wings of this species, and how it is limited to males, when both sexes share practically the same genome.
Watch how model organisms are used at the IBDM to investigate normal and derailed stem cell trajectories.
Sweet Temptations: How Drosophila Suzukii’s Love for Sugar is Causing Havoc in the Agriculture Industry.
Seeking for your Master internship? The IBDM seems like the right place to do it? Check out our offers.
A publication from the Prud’homme team shows how the sexually dimorphic expression pattern of an X-linked gene is controlled by an interallelic interaction, a phenomenon known as transvection.
Benoit Aigouy, from the Prud’homme team, developed EPySeg, an open-source software that uses deep learning to segment membrane-stained epithelial tissues, from any organism, automatically and very efficiently.
A publication from the Prud’homme team, in collaboration with the group of N. Gompel explores how the spatial pattern and quantitative regulatory information is encoded in the enhancer sequence of a developmental gene.
During the Master’s project (M1 or M2), the student will use some of the techniques used in the laboratory to quantitatively interrogate the structure-function relationship underlying interallelic interactions.


To study how animal shapes are encoded in the genomic sequence, how they emerge during development, and how they appear during evolution, we mostly focus on the formation and evolution of insect pigmentation patterns. These patterns are perfect morphological readout of the expression patterns of the genes underlying their formation. Through the analysis of the specification and the evolution of pigmentation patterns, we, therefore, address general questions about how gene expression patterns are established and how they evolve.
We are studying a particular wing pigmentation pattern, the wing spot, that has evolved repeatedly in several evolutionary lineages of Drosophila species (Prud’homme et al., Nature 2006). We focus on the regulation of a gene named yellow, which is essential for the production of black pigment in all insects. In the male wing of Drosophila wing-spotted species, yellow has evolved a spot-like expression pattern. This novel expression pattern is driven by a ‘spot’ enhancer, which has evolved by co-opting a nearby ‘wing’ enhancer, active throughout the wing, and present in all Drosophila species (Gompel, et al., Nature 2005).
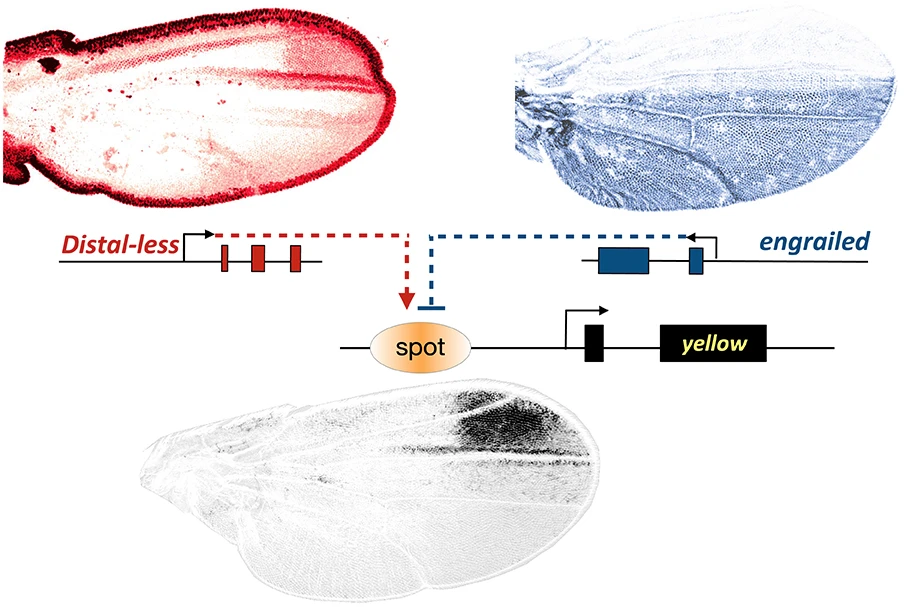
We have found that the evolutionary-derived yellow spot enhancer activity has evolved by recruiting, in particular, two deeply conserved transcription factors essential to the making of a wing, Engrailed and Distal-less (Gompel, et al., Nature 2005; Arnoult et al., Science 2013). The evolution of regulatory links between these transcription factors, and others, and the enhancers of pigmentation genes like yellow were at the origin of a novel wing pigmentation pattern.
Subsequently, through the diversification of the evolutionary lineage, the wing spot pattern changed shape, driven by changes in the regulation of the top regulator Distal-less (Arnoult et al., Science 2013).

The genetic principles of pigmentation pattern formation that we identified by studying the wing of fruit flies are also valid in other organisms. Indeed, we have shown that the variation in pigmentation patterns on the elytra of the ladybird Harmonia axyridis, for which more than 200 color morphs are known in natural populations, is fueled by the same type of regulatory principles we have uncovered in fruit flies (Gautier et al., Current Biology 2018).
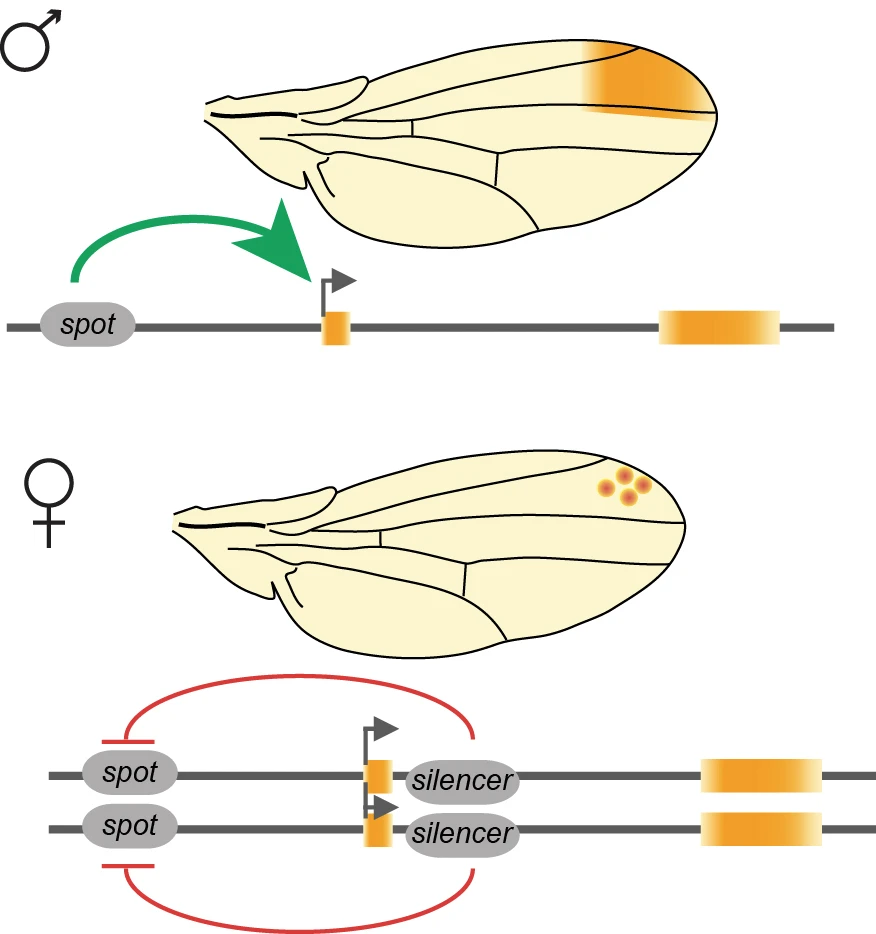
The wing spot is present only in males, and the yellow gene, accordingly, is expressed differently in the wing of the two sexes. We found that this sexually dimorphic regulation of yellow is not controlled by the canonical sex determination pathway that controls all differences between sexes in Drosophila. Instead, we found that the sexually dimorphic regulation of the spot enhancer activity is controlled by transvection (Galouzis & Prud’homme, Science 2021). Transvection, or proximity-dependent interallelic interaction, is a regulatory mechanism that relies on interchromosomal communication. Since yellow is on the X chromosome it is present in two copies in females, but only one in males. Therefore, the yellow alleles can only interact in females, which leads to the silencing of the spot enhancer.
We are currently addressing the mechanisms of transvection at the yellow locus by studying how the yellow alleles find each other in the nuclear space; what is the minimal distance required for their functional interaction; and how the physical proximity of the two alleles leads to the silencing of the spot enhancer. For this, we combine molecular and genetics approaches with quantitative microscopy methods. Our long-term goal is to better understand the relationship between the genome topology, in particular the homologous chromosomes, and the regulation of gene expression.

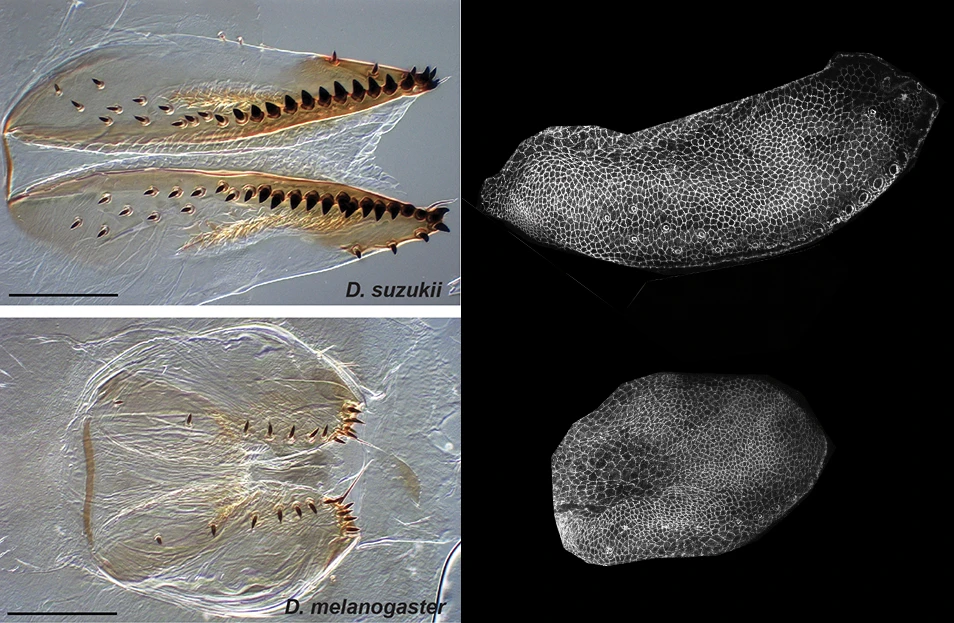
D. suzukii’s egg-laying organ, or ovipositor, has evolved a particular size and shape that allow piercing through the skin of ripe fruit. We have shown that the morphological changes of the ovipositor evolved mostly in a short time window during pupal development. The evolutionary modification of the ovipositor morphogenesis relies on the fine-tuning of cell size expansion and re-organization of the tissue (Green et al, Current Biology 2019).
We are currently extending these observations on fixed tissues with comparative live-imaging of the ovipositor morphogenesis. To facilitate the study of epithelial tissue morphogenesis (tracking the cells and measuring the transformation of the tissue) we have developed an imaging analysis software that uses deep learning to automatically segment cell contours (Aigougy et al., Development 2020).
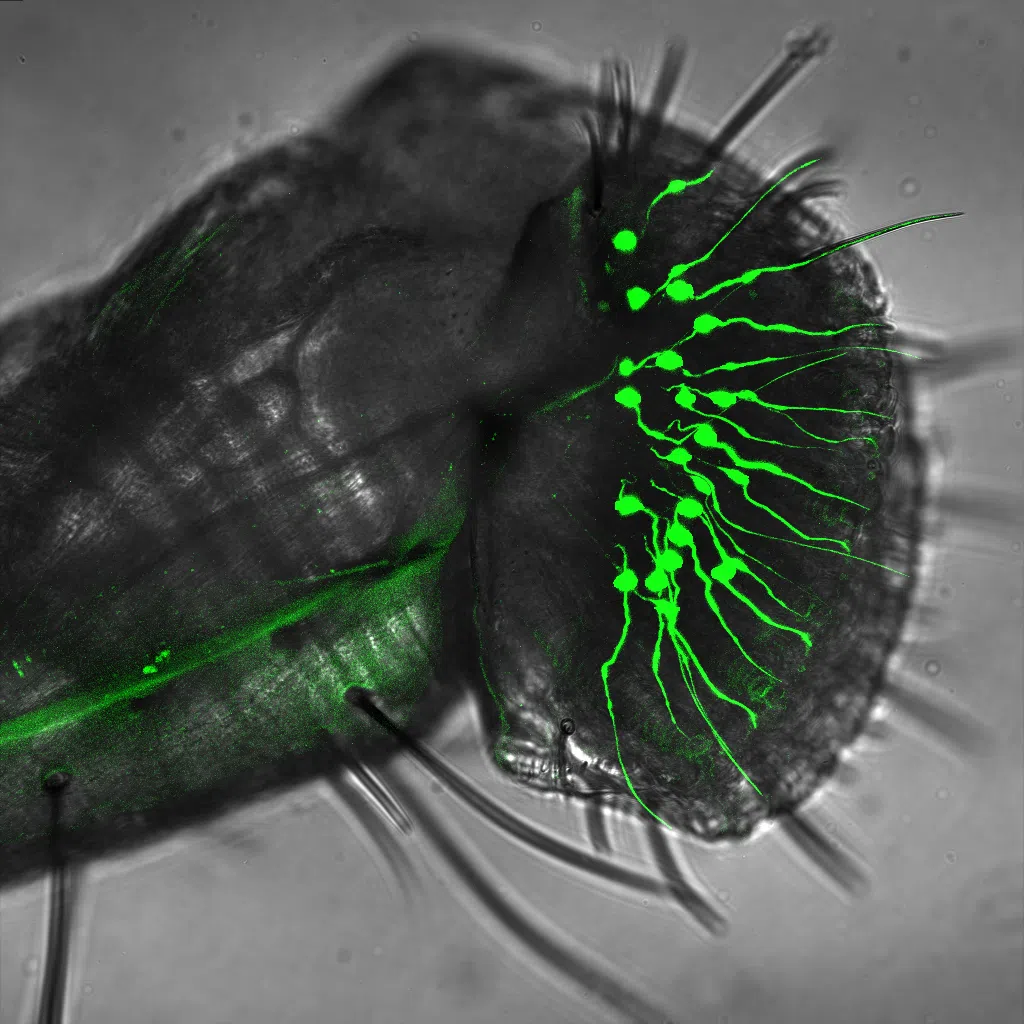
In addition to the modification to the ovipositor, which grants access to a novel ecological niche to D. suzukii, the egg-laying site choice of this species has also changed compared to other closely related Drosophila species. We have established that several sensory modalities, including olfaction and gustation, have changed in D. suzukii to drive its novel egg-laying behavior.
We have developed targeted mutagenesis and functional transgenesis protocols to study the egg-laying behavior of D. suzukii (Karageorgi et al., Current Biology 2017), and we have established a near-chromosome level assembly and annotation of D. suzukii’s genome (Paris et al., Scientific Reports 2020).
Our goal is to identify the olfactory and gustatory cues, and the neural circuits, that drive the egg-laying preference evolutionary shift in D. suzukii. To this end, we are using a combination of behavioral assays, genetic manipulations, and functional imaging in D. suzukii and D. melanogaster.