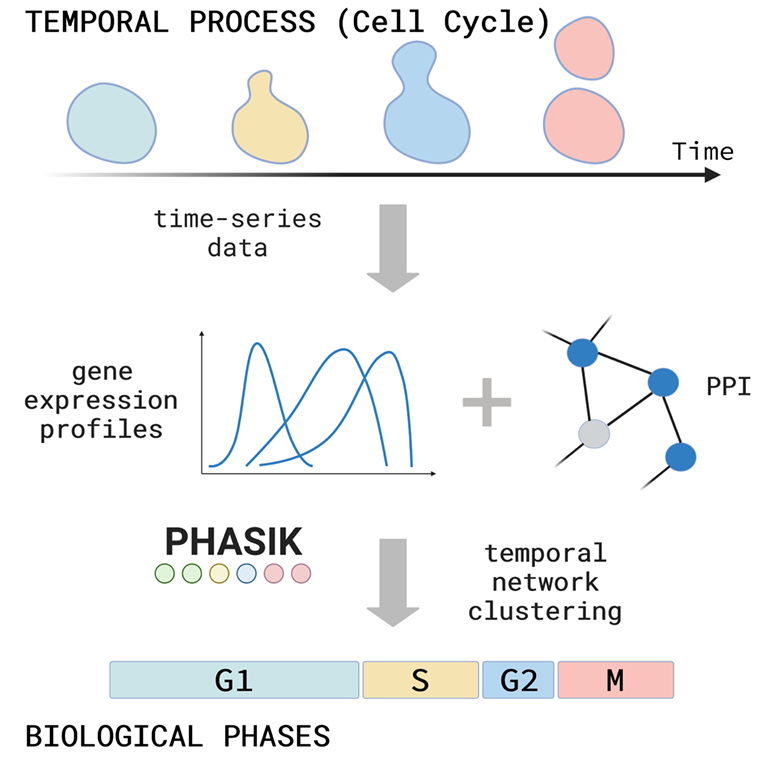
Many biological systems are organised in temporal phases or states. The cell cycle, for example, has 4 major phases: a first Gap phase (G1), in which the cell starts to grow and determines if it should divide or not, followed by a synthesis phase (S) in which DNA is replicated, then another Gap phase (G2), in which the cell further grows and prepares for the final phase, Mitosis (M), in which the duplicated genome is separated in two daughter cells. Circadian rhythm is a second example, in which key clock genes regulate gene expression in a daily 24-hour rhythm that has evolved to adapt to the daily light and dark cycles. Such temporal phases or states of a biological system are created by a precisely timed availability of specific regulatory proteins in a cell, as well as their precise interactions with one another. These changes in the states of the cells or biological systems can be measured by quantifying the levels of protein or mRNA (as an approximation) in time-series experiments.
We developed a method, Phasik, to extract phase information from these time-series experiments by combining temporal expression data with interaction data from protein interaction networks. We build a (partially) temporal network and then cluster the ‘snapshots’ of this temporal network. We extensively tested Phasik and could show that we can reliably recover the well-known phases and sub-phases of the cell cycle and detect cell cycle arrest in mutants. With Phasik, we can also recover the 24-hour rhythm of the circadian clock and we show that this rhythm is perturbed in clock-gene mutants.
Phasik is the first algorithm for temporal network clustering applied and available for biological systems. Moreover, it is the very first time that we demonstrate that partial temporal information for just a subset of interactions in the network is sufficient for extracting phases from a temporal system. Phasik can be used to infer phases for a wide range of less well-known biological systems and can help gain insight in their underlying mechanisms. The code is freely available as a Python package with Jupyter notebooks.
To know more
Inferring cell cycle phases from a partially temporal network of protein interactions
Authors: Maxime Lucas (IBDM, CPT, I2M), Arthur Morris (University of Oxford), Alex Townsend-Teague (University of Oxford), Laurent Tichit (I2M), Bianca Habermann (IBDM), Alain Barrat (CPT)
Funding: The Turing Centre for Living Systems (CENTURI, ANR-16-CONV-0001)