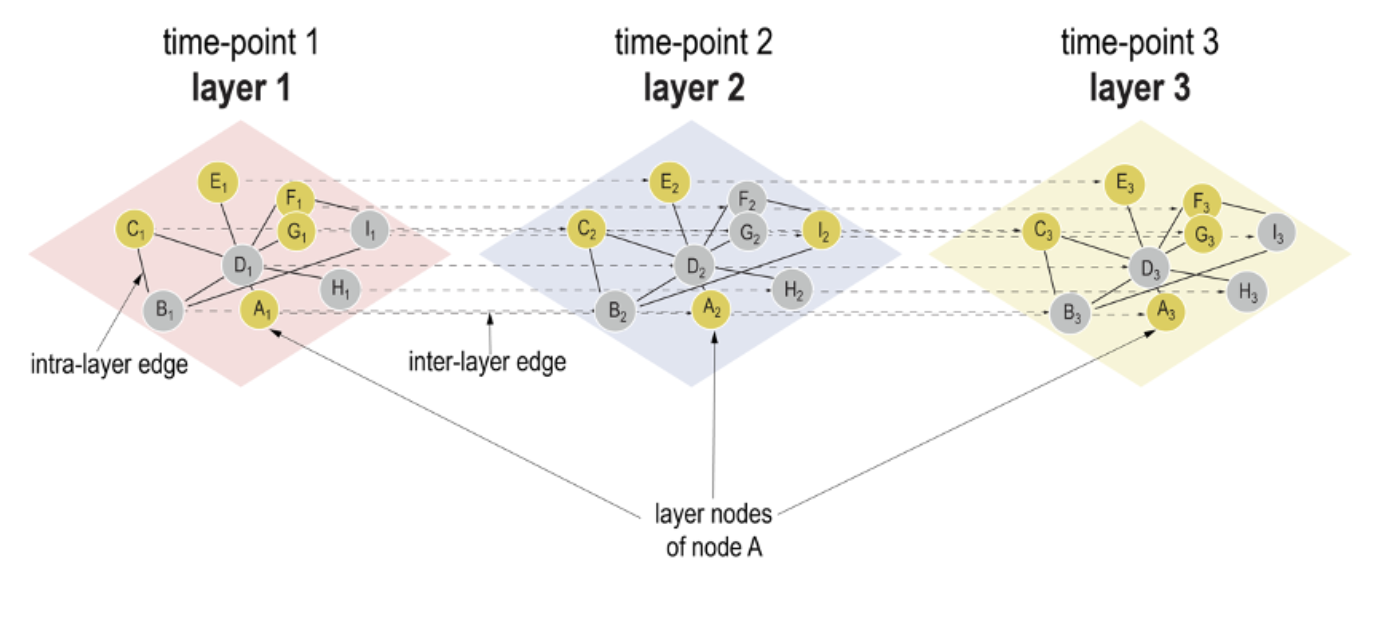
The temporal resolution of a system is important. We can measure that during cyclic processes such as the cell cycle, where genes are up- or down-regulated at certain time-points of the cycle, or during development or disease onset, where genes are induced at certain time-points in a tissue. Tools to analyze such temporal data, identifying for instance temporally active pathways, are largely lacking. With TimeNexus, a new Cytoscape app is introduced that allows you to integrate temporal expression data, e.g. coming from a time-series RNA-seq experiment, with a protein interaction network, and to look for pathways that are deregulated over time. To achieve this, the Habermann team has repurposed the concept of multilayer networks generally used to integrate different types of data, e.g. from a transcriptomic and proteomic screen. TimeNexus places the data from consecutive time points on consecutive layers of the multilayer network, generating a temporal multilayer network (tMLN). By applying some little tricks and ‘flattening’ this tMLN, TimeNexus can then use available Cytoscape apps to look for pathways that contain deregulated genes over time. You can download the app from the Cytoscape app store. This work was the PhD-project of Michael Pierrelee and the result of a successful collaboration between the Habermann and Moqrich teams, together with Laurent Tichit from I2M and Fabrice Lopez from TAGC.
Find out more: Pierrelée M, Reynders A, Lopez F, Moqrich A, Tichit L, Habermann BH. Introducing the novel Cytoscape app TimeNexus to analyze time-series data using temporal MultiLayer Networks (tMLNs). Sci Rep. 2021 Jul 1;11(1):13691. doi: 10.1038/s41598-021-93128-5.