Congenital heart defects occur in 1 in 100 births and approximately one third of these affect the cardiac outflow tract, where the great arteries, the aorta and pulmonary trunk, align with the left and right ventricles respectively. Myocardium at the base of the aorta and pulmonary trunk has been shown to originate in distinct regions of the unseptated outflow tract at midgestation and to be prepatterned in the progenitor field in the early embryo, from which cells add progressively to the growing heart tube.
Genetic or environmental perturbation of progenitor cell deployment leads to outflow tract CHD in man and mouse models. For example, the 22q11.2 deletion (also known as DiGeorge) syndrome gene, encoding the transcription factor TBX1, is expressed in future sub-pulmonary myocardium and required for correct addition of these progenitor cells to the heart.
Starting from an analysis of genes differentially expressed in the outflow tract, Rammah and colleagues in the Kelly group “Genetic control of heart development” have used in vitro and in vivo approaches to identify the lipid sensor PPARg as a Tbx1-dependent regulator of subpulmonary myocardial development. PPARg expression is repressed in future sub-aortic myocardium by the Notch signaling pathway, through the activities of the transcription factor HES1 and the non-canonical Notch ligand DLK1, a negative regulator of PPARg during adipogenesis.
These findings reveal how complementary regulatory cascades establish regional identity in the developing outflow tract and prefigure later septation events. Moreover, these results point to the importance of metabolic changes and the gene-environment interface during heart development.
The perspectives raised by this work are being followed up in the Pp-heart ANR project in collaboration with Magali Théveniau-Ruissy and Francesca Rochais in the Marseille Medical Genetics Unit, Aix Marseille University.
To know more
PPARγ and NOTCH Regulate Regional Identity in the Murine Cardiac Outflow Tract.
Rammah M, Théveniau-Ruissy M, Sturny R, Rochais F, Kelly RG.
Circ Res. 2022 Oct 7:101161CIRCRESAHA122320766.
doi: 10.1161/CIRCRESAHA.122.320766. Online ahead of print.
PMID: 362051
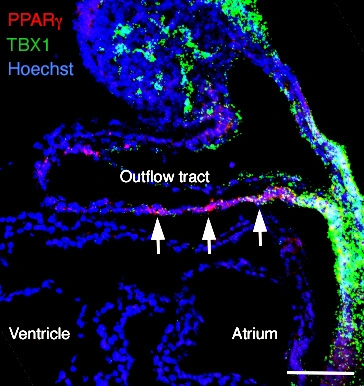
Figure legend
The figure shows regionalised Pparg transcript accumulation (red) in the inferior wall of the midgestation outflow tract and Tbx1 (green) in adjacent cardiac progenitor cells. Sagittal section of an embryonic day 9.5 mouse embryo; scale bar: 100 μm. Photo: Magali Théveniau-Ruissy.