Computer Science, Morphogenesis and Variability
Graph Theory, Machine Learning, and Big Data drive our lab and our analyses of morphogenesis and developmental reproducibility across species from fluorescent microscopy to spatial omics acquisitions.
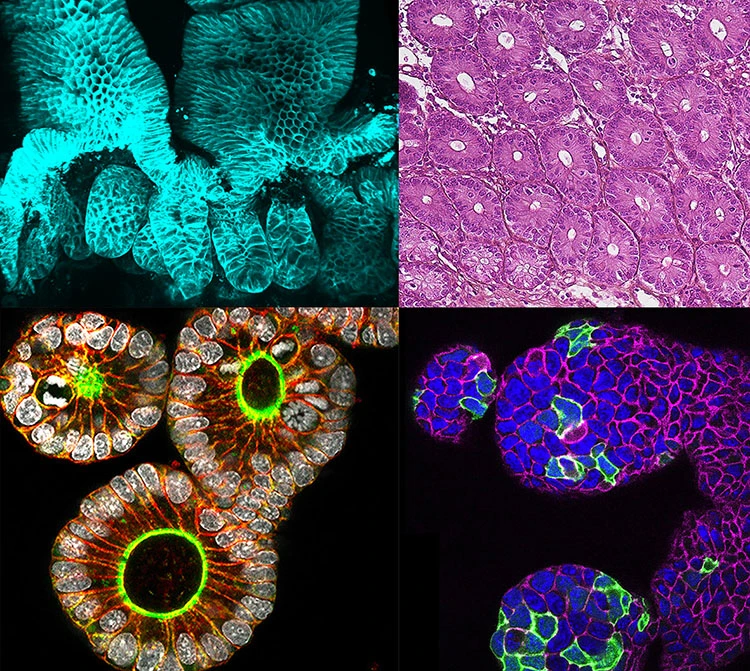
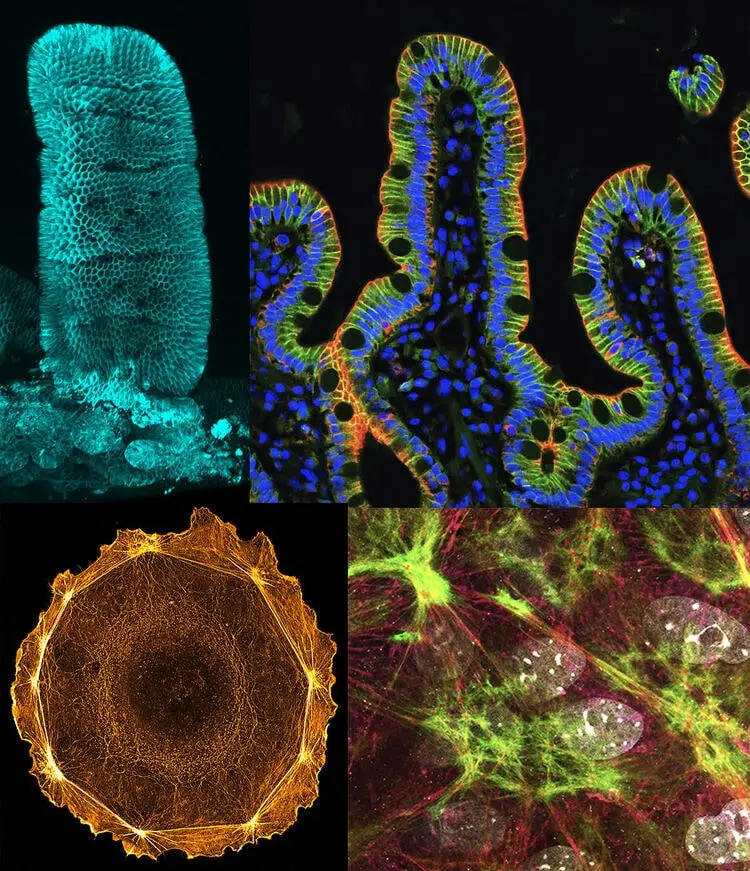
Our lab operates at the interface of computer science and developmental biology. We focus on the development of computational methods for analysing morphogenesis at the single-cell scale in whole organisms throughout their development.
Our primary objective is to apply computer vision, graph-based, machine learning and big data algorithms to quantify and comprehend developmental reproducibility during embryogenesis in various model organisms.
Our ultimate aim is to investigate the impact of developmental variability on morphogenesis by utilising homemade statistical averages from two main data modalities: fluorescence microscopy images and spatial omics.
Our research revolves around three key areas:
- Computer Vision: We are engaged in developing computer vision and machine learning algorithms to reconstruct and analyse average representations of developing organisms using 3D fluorescence microscopy time-series data. This enables us to identify trends and patterns in developmental processes.
- Spatial Omics Data: We also delve into graph analysis, data structures, and visualisation tools to effectively manage and represent complex 3D spatial omics datasets. This aids in uncovering the molecular aspects of developmental processes.
- Average Integration of Modalities: Our core strength lies in integrating both fluorescence microscopy images and spatial omics data into comprehensive average atlases of embryogenesis across multiple species.These atlases provide valuable insights into both shared and unique aspects of development within and across species.
Our lab maintains collaborative partnerships with developmental biologists and physicists in Marseille, Paris (France), Heraklion (Greece), Cleveland (Ohio, USA), New York (New York, USA), and other regions. Through these collaborations, we aim to enhance our understanding of fundamental biological processes while promoting interdisciplinary research.
Publications
Spatiotemporal transcriptomic maps of whole mouse embryos at the onset of organogenesis
Mathematical and bioinformatic tools for cell tracking
Contact area-dependent cell communication and the morphological invariance of ascidian embryogenesis
Spatiotemporal transcriptomic maps of whole mouse embryos at the onset of organogenesis
Mathematical and bioinformatic tools for cell tracking
Contact area-dependent cell communication and the morphological invariance of ascidian embryogenesis
News
IBDM welcomes a new research group!
We are delighted to welcome Leo Guignard and his new research group ‘Computer Science, Morphogenesis and Variability’ to IBDM.
No jobs opportunities found..