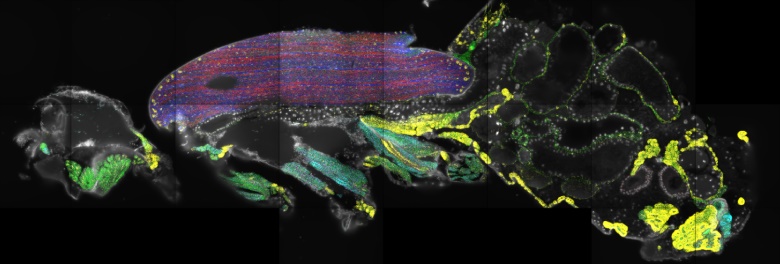
Spatial transcriptomics documents differential mRNA expression patterns in different adult muscle types. Note the high expression of fly titin (large yellow dots) in the nuclei close to the ends of the large flight muscles (in blue/red).
The fruit fly Drosophila has been a simple model for genetics, developmental biology, and behaviour for over 100 years. The Drosophila genome contains about 13,000 genes expressed in many cell types. Researchers used DNA-sequencing methods to catalogue the expression of mRNAs in thousands of cells isolated from the adult fly. This led to the Fly Cell Atlas, defining several 100 different cell types, which differ by the combination of active genes.
Many of these cell types are located in the fly brain, but the exact position of several cell types was unknown, because the sequencing method used isolated cells. In a collaboration between the Aerts group in Leuven and the Schnorrer group in Marseille, we have now determined the location of a number of these unknown cell types in the fly brain and also studied the expression of mRNAs in the fly body using a new method called spatial transcriptomics. This method determines the positions where a gene is active in the fly body. We used sections of the adult head to measure expression in the brain, and sections of the adult body to determine expression in the entire body for a total of 150 different genes.
Spatial transcriptomics allowed us to determine the location of all the major cell types of the adult. It also identified the location of formerly unknown cell types in the fly brain, whose existence had only been defined from the Fly Cell Atlas data. Unexpectedly, we found that in large muscle cells, which contain 100s of nuclei, some mRNAs are preferentially produced from a small subset of nuclei. This suggests that not all nuclei of a muscle cell follow the same transcriptional program at a given time. In the future, it will be interesting to discover the mechanism of how this can be achieved.
Furthermore, the protocols and computational methods presented in this study show how to combine cell atlas data with spatial transcriptomics to determine where identified cells are located in the body of an organism.
Janssens J, Mangeol P, Hecker N, Partel G, Spanier KI, Ismail JN, Hulselmans GJ, Aerts S, Schnorrer F. Spatial transcriptomics in the adult Drosophila brain and body. Elife. 2025 Mar 18;13:RP92618.